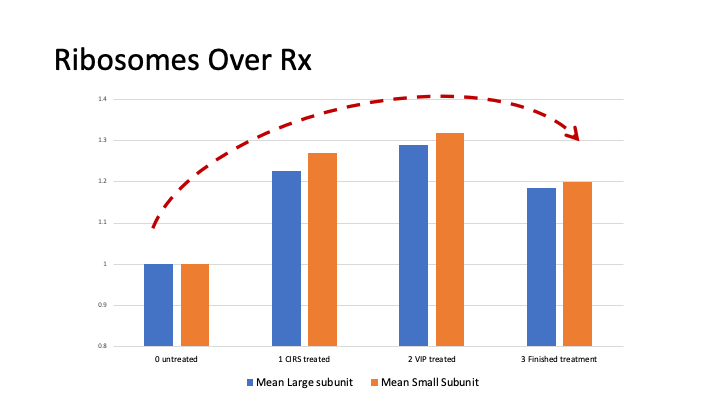
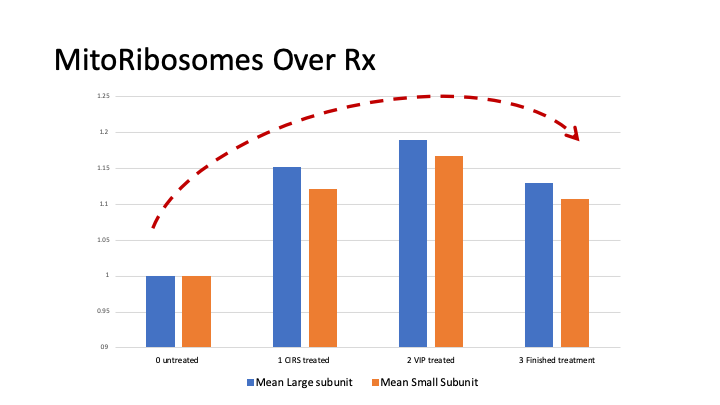
Progene DX has identified statistically significant, differentially expressed, genes and proteins in the blood of CIRS patients that now has been leveraged to produce better tools and tests for diagnosis and case management. Not only will these test identify CIRS in new patients, they can also be used to track recovery throughout the course of treatment. This road to recovery is not straight forward, in fact its curved, what we call the CIRS curve (see below). As the body comes out of the winter of CIRS, it goes through a predictable progression of systems that get turned on once again. We can monitor this progression with our GENIE assay.
Gene expression measurements are taken by extracting total ribonucleic acid (total RNA) from peripheral blood cells (PBLs) and running a limited (187 gene) transcriptomic analysis in our Boston laboratory. The results of the GENIE gene expression test are best interpreted when accompanied with blood protein biomarkers of CIRS, click HERE for the full panel of protein tests. These blood protein labs can be acquired through major national clinical diagnostic labs like Quest or LabCorp.
However, Progene DX will soon be opening a new protein testing facility alongside its gene expression lab just outside of Boston.
Proteomics
Objective diagnosis of CIRS is a critical first step in applying restorative therapies. Many irregularities in protein expression have now been identified in patients with CIRS, dominated by (1) lack of regulation of host inflammatory response as evidenced by deficiency of alpha melanocyte stimulating hormone (MSH) and/or vasoactive intestinal polypeptide (VIP); (2) presence of more than one of Th1 responses (pro-inflammatory); Th2 responses (anti-inflammatory); Th17 responses (tied to transforming growth factor beta-1 (TGFβ-1)); coagulation abnormalities, especially abnormalities in von Willebrand’s profile; activation of complement split products; activation of elements under regulation of hypoxia inducible factor including vascular endothelia growth factor (VEGF) and erythropoietin; abnormal regulation of ACTH responses to cortisol and ADH responses to osmolality.
Genomics
The genomics investigation of CIRS is focused on the expression of genes, or transcriptomics. Our research has identified nearly 2,000 statistically significant, differentially expressed genes in the average CIRS patient before they begin CIRS treatment. These abnormalities are dominated by genes involved in ribosomal protein and mitochondrial activity, resulting in a hypo metabolic cellular state. Additionally, many patients exhibit abnormalities in T cell receptors, insulin signaling, coagulation and various CD markers. We also looked at micro RNAs and have identified that Let7 and MIR23 family members contribute to CIRS pathology.